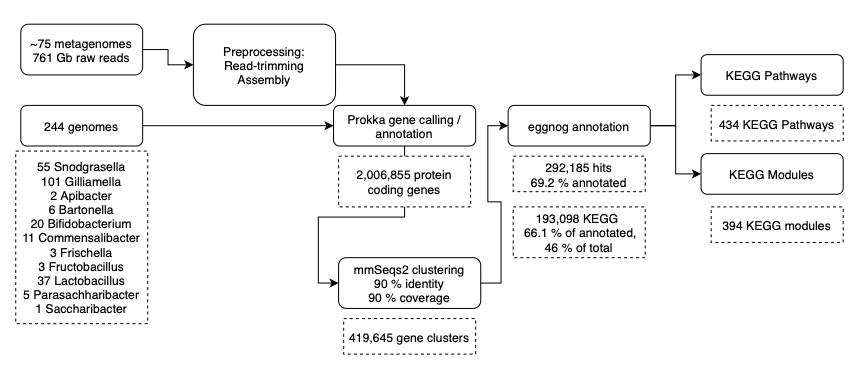
The Plan
In this project I will use publically available data to
- estimate the functional composition of a “normal” bee gut microbiome
- assemble in silico communties of bee gut strains
- estimate how well designed communities recapitulate natural microbiomes
Datasets
I used three published sets of metagenomes from:
- T. Regan, M. W. Barnett, D. R. Laetsch, S. J. Bush, D. Wragg, G. E. Budge, F. Highet, B. Dainat, J. R. de Miranda, M. Watson, M. Blaxter, T. C. Freeman, Characterisation of the British honey bee metagenome. Nat. Commun. 9, 4995 (2018).
- P. Engel, V. G. Martinson, N. A. Moran, Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. U. S. A. 109, 11002–11007 (2012).
- K. M. Ellegaard, P. Engel, Genomic diversity landscape of the honey bee gut microbiota. Nature Communications. 10 (2019).
I downloaded isolate genomes from the NCBI Assembly database, computationally annotated some metadata through entrez, and then manually added additional metadata (host species).